High throughput chemical methods were to be used to make large numbers of predicted proteins and protein domains, based on microbial genome sequences. excellence, Structure To browse Academia.edu and the wider internet faster and more securely, please take a few seconds toupgrade your browser. 3, Issue 1, Proceedings of the National Academy of Sciences, Vol. for Research on Aging, Novato, CA (United States), Univ. Bioinformatics Group, Purdue Univ., West Lafayette, IN (United States). Dept. P. Radivojac, W.T. DomainSVM method utilizes evidence of multiple interacting domains to predict a protein interaction. of California, San Francisco, CA (United States). Conclusions: The top-performing methods in CAFA2 outperformed those from CAFA1. Inst. Here we present DomainSVM, a predictive method for PPI that uses computationally inferred domain-domain interaction values in a Support Vector Machine framework to predict protein interactions. 28, Issue 12, PLoS Computational Biology, Vol. facilities. of Bioengineering and Therapeutic Sciences, Colorado State Univ., Fort Collins, CO (United States). Fifty-four methods representing the state of the art for protein function prediction were evaluated on a target set of 866 proteins from 11 organisms. Gene Ontology: tool for the unification of biology. Intrinsically disordered proteins and intrinsically disordered protein regions. Dept. Oron, A.M. Schnoes, T. Wittkop, Y.A.I. Division of Molecular Biosciences; Spanish National Research Council (CSIC), Madrid (Spain). of California, Berkeley, CA (United States). 295, Issue 2, Journal of Aquatic Food Product Technology, Vol. Univ.
44, Issue 14, Royal Society Open Science, Vol. Fenner, Kathrin; Canonica, Silvio; Wackett, Lawrence P. Hornung, Bastian; Martins dos Santos, Vitor A. P.; Smidt, Hauke, Yu, Guoxian; Zhu, Hailong; Domeniconi, Carlotta, Callahan, Alison; Cifuentes, Juan Jos; Dumontier, Michel, Lena, Pietro Di; Domeniconi, Giacomo; Margara, Luciano, vek, Clemens; Friedrichs, Gerald; Heizinger, Leonhard, Alborzi, Seyed Ziaeddin; Ritchie, David W.; Devignes, Marie-Dominique, Teso, Stefano; Masera, Luca; Diligenti, Michelangelo, Perlasca, Paolo; Frasca, Marco; Ba, Cheick Tidiane. 9, Issue 5, PLOS Computational Biology, Vol. Hoehndorf, R.; Schofield, P. N.; Gkoutos, G. V. Koskinen, Patrik; Trnen, Petri; Nokso-Koivisto, Jussi, Alshahrani, Mona; Khan, Mohammad Asif; Maddouri, Omar, Mahlich, Yannick; Steinegger, Martin; Rost, Burkhard, Khan, Ishita K.; Jain, Aashish; Rawi, Reda, Morandin, Claire; Havukainen, Heli; Kulmuni, Jonna, Motion, Graham B.; Howden, AndrewJ.M.; Huitema, Edgar, Tatusova, Tatiana; DiCuccio, Michael; Badretdin, Azat, Zhu, Chengsheng; Miller, Maximilian; Marpaka, Srinayani. of Missouri, Columbia, MO (United States). Computational Bioscience Program. 284, Issue 31, Molecular Systems Biology, Vol. of Padova (Italy). Here we report the results from the first large-scale community-based critical assessment of protein function annotation (CAFA) experiment. 08, Issue 02, PLoS Computational Biology, Vol. campus tour, Connect with A large-scale evaluation of computational protein function prediction, Mathematical and Statistical Methods - Biometris, Nature Methods : techniques for life scientists and chemists. of Electrical Engineering and Computer Science. and governance, Alumni & sections dedicated to protein function research, with an emphasis on the theory and practice of computational methods utilized in functional annotation. Dive into the research topics of 'A large-scale evaluation of computational protein function prediction'. Wimalanathan, Kokulapalan; Lawrence-Dill, Carolyn J. Vidulin, Vedrana; muc, Tomislav; Deroski, Sao. Ashburner, Michael; Ball, Catherine A.; Blake, Judith A. Kourmpetis, Yiannis A. I.; van Dijk, Aalt D. J.; Bink, Marco C. A. M. Wang, Dennis Ding-Hwa; Shu, Zhanyong; Lieser, Scot A. Vazquez, Alexei; Flammini, Alessandro; Maritan, Amos, Sharan, Roded; Ulitsky, Igor; Shamir, Ron.
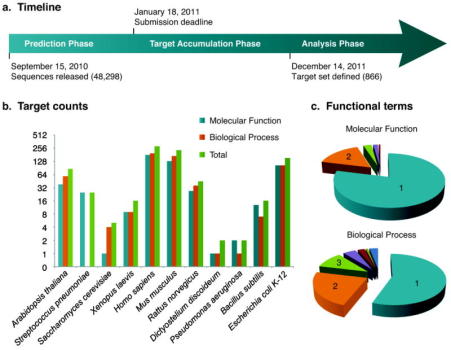 journal = "Nature Methods : techniques for life scientists and chemists", Radivojac, P, Clark, WT, Oron, TR, Schnoes, AM, Wittkop, T, Kourmpetis, YAI. The meeting was exciting and, based on feedback, quite successful. 38, Issue 1, Journal of Evolutionary Biology, Vol. College of Medicine. 31, Issue 8, Nucleic Acids Research, Vol. Such patterns suggest relationships in the data that only a biology domain expert might be able to explain. School of Medicine.
journal = "Nature Methods : techniques for life scientists and chemists", Radivojac, P, Clark, WT, Oron, TR, Schnoes, AM, Wittkop, T, Kourmpetis, YAI. The meeting was exciting and, based on feedback, quite successful. 38, Issue 1, Journal of Evolutionary Biology, Vol. College of Medicine. 31, Issue 8, Nucleic Acids Research, Vol. Such patterns suggest relationships in the data that only a biology domain expert might be able to explain. School of Medicine. of Plant and Microbial Biology; Wellcome Trust Genome Campus, Hinxton (United Kingdom). Centre for Systems and Synthetic Biology. There were 73 registered delegates at the meeting. Have you forgotten your guest credentials? Two findings stand out: (i) today's best protein function prediction algorithms substantially outperform widely used first-generation methods, with large gains on all types of targets; and (ii) although the top methods perform well enough to guide experiments, there is considerable need for improvement of currently available tools.". of Plant and Microbial Biology, Miami Univ., Oxford, OH (United States). Biometris; Nestl Inst. https://doi.org/10.1148/radiology.143.1.7063747, https://doi.org/10.1007/s00018-003-3114-8, PNPASE Regulates RNA Import into Mitochondria, https://doi.org/10.1016/j.cell.2010.06.035, https://doi.org/10.1016/S0168-9525(99)01706-0, Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles, Predicting protein function from protein/protein interaction data: a probabilistic approach, https://doi.org/10.1093/bioinformatics/btg1026, Down-regulation of Myc as a Potential Target for Growth Arrest Induced by Human Polynucleotide Phosphorylase (hPNPase) in Human Melanoma Cells, Gene networks in Drosophila melanogaster: integrating experimental data to predict gene function, Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, https://doi.org/10.1016/S0968-0004(98)01335-8, The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata, Gene Ontology: tool for the unification of biology, Bayesian Markov Random Field Analysis for Protein Function Prediction Based on Network Data, https://doi.org/10.1371/journal.pone.0009293, Human Mitochondrial SUV3 and Polynucleotide Phosphorylase Form a 330-kDa Heteropentamer to Cooperatively Degrade Double-stranded RNA with a 3-to-5 Directionality, ConFuncfunctional annotation in the twilight zone, https://doi.org/10.1093/bioinformatics/btn037, A scalable method for integration and functional analysis of multiple microarray datasets, https://doi.org/10.1093/bioinformatics/btl492, Global protein function prediction from protein-protein interaction networks, Networkbased prediction of protein function, Computational Methods for Identification of Functional Residues in Protein Structures, https://doi.org/10.2174/138920311796957685, Prediction of Human Protein Function from Post-translational Modifications and Localization Features, https://doi.org/10.1016/S0022-2836(02)00379-0, Analysis of the human polynucleotide phosphorylase (PNPase) reveals differences in RNA binding and response to phosphate compared to its bacterial and chloroplast counterparts, Detecting Protein Function and Protein-Protein Interactions from Genome Sequences, https://doi.org/10.1126/science.285.5428.751, Inference of Protein Function from Protein Structure, https://doi.org/10.1016/j.str.2004.10.015, Enhanced automated function prediction using distantly related sequences and contextual association by PFP, Testing the Ortholog Conjecture with Comparative Functional Genomic Data from Mammals, https://doi.org/10.1371/journal.pcbi.1002073, Analysis of protein function and its prediction from amino acid sequence, Whole-proteome prediction of protein function via graph-theoretic analysis of interaction maps, https://doi.org/10.1093/bioinformatics/bti1054, Protein Molecular Function Prediction by Bayesian Phylogenomics, https://doi.org/10.1371/journal.pcbi.0010045, A Probabilistic Functional Network of Yeast Genes, Domain-Based and Family-Specific Sequence Identity Thresholds Increase the Levels of Reliable Protein Function Transfer, https://doi.org/10.1016/j.jmb.2008.12.045, Protein function prediction the power of multiplicity, https://doi.org/10.1016/j.tibtech.2009.01.002, Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium, The GOA database in 2009--an integrated Gene Ontology Annotation resource, HIERARCHICAL CLASSIFICATION OF GENE ONTOLOGY TERMS USING THE GOstruct METHOD, https://doi.org/10.1142/S0219720010004744, The Rough Guide to In Silico Function Prediction, or How To Use Sequence and Structure Information To Predict Protein Function, https://doi.org/10.1371/journal.pcbi.1000160, Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress, https://doi.org/10.1016/j.bbrc.2008.05.058, Predicting function: from genes to genomes and back 1 1Edited by P. E. Wright, Prediction of Protein Function Using ProteinProtein Interaction Data, https://doi.org/10.1089/106652703322756168, Enzyme Promiscuity: A Mechanistic and Evolutionary Perspective, https://doi.org/10.1146/annurev-biochem-030409-143718, Automated prediction of protein function and detection of functional sites from structure, Annotation Error in Public Databases: Misannotation of Molecular Function in Enzyme Superfamilies, https://doi.org/10.1371/journal.pcbi.1000605, Accuracy of two pulse-oximetry measurements for INTELLiVENT-ASV in mechanically ventilated patients: a prospective observational study, https://doi.org/10.1038/s41598-021-88608-7, https://doi.org/10.1186/1465-6906-11-s1-p21, GOtcha: a new method for prediction of protein function assessed by the annotation of seven genomes, https://doi.org/10.6084/m9.figshare.2837.v2, Protein function in precision medicine: deep understanding with machine learning, Evolutionary couplings and sequence variation effect predict protein binding sites, Structural disorder in plant proteins: where plasticity meets sessility, https://doi.org/10.1007/s00018-017-2557-2, Beyond Homology Transfer: Deep Learning for Automated Annotation of Proteins, https://doi.org/10.1007/s10723-018-9450-6, Insect-associated bacterial communities in an alpine stream, https://doi.org/10.1007/s10750-019-04097-w, Construction of a synthetic protein using PCR with a high essential amino acid content for nutritional purposes, https://doi.org/10.1007/s11033-019-04604-1, The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera, Network enhancement as a general method to denoise weighted biological networks, https://doi.org/10.1038/s41467-018-05469-x, YesU from Bacillus subtilis preferentially binds fucosylated glycans, https://doi.org/10.1038/s41598-018-31241-8, GOGO: An improved algorithm to measure the semantic similarity between gene ontology terms, https://doi.org/10.1038/s41598-018-33219-y, A postprocessing method in the HMC framework for predicting gene function based on biological instrumental data, Panoramic view of a superfamily of phosphatases through substrate profiling, Complex evolutionary footprints revealed in an analysis of reused protein segments of diverse lengths, Topology-function conservation in proteinprotein interaction networks, https://doi.org/10.1093/bioinformatics/btv026, Exploiting ontology graph for predicting sparsely annotated gene function, https://doi.org/10.1093/bioinformatics/btv260, POSSUM: a bioinformatics toolkit for generating numerical sequence feature descriptors based on PSSM profiles, https://doi.org/10.1093/bioinformatics/btx302, Improving protein function prediction using protein sequence and GO-term similarities, https://doi.org/10.1093/bioinformatics/bty751, Robust Method for Detecting Convergent Shifts in Evolutionary Rates, HAMAP in 2015: updates to the protein family classification and annotation system, PredictProteinan open resource for online prediction of protein structural and functional features, Gene3D: expanding the utility of domain assignments, APRICOT: an integrated computational pipeline for the sequence-based identification and characterization of RNA-binding proteins, COFACTOR: improved protein function prediction by combining structure, sequence and proteinprotein interaction information, CATH: expanding the horizons of structure-based functional annotations for genome sequences, INGA 2.0: improving protein function prediction for the dark proteome, NetGO: improving large-scale protein function prediction with massive network information, Identifying gene function and module connections by the integration of multispecies expression compendia, Interactions in the microbiome: communities of organisms and communities of genes, Temperature- and sex-related effects of serine protease alleles on larval development in the Glanville fritillary butterfly, Evaluating Pesticide Degradation in the Environment: Blind Spots and Emerging Opportunities, Studying microbial functionality within the gut ecosystem by systems biology, https://doi.org/10.1186/s12263-018-0594-6, Predicting protein functions using incomplete hierarchical labels, https://doi.org/10.1186/s12859-014-0430-y, An evidence-based approach to identify aging-related genes in Caenorhabditis elegans, https://doi.org/10.1186/s12859-015-0469-4, GOTA: GO term annotation of biomedical literature, https://doi.org/10.1186/s12859-015-0777-8, An assessment of catalytic residue 3D ensembles for the prediction of enzyme function, https://doi.org/10.1186/s12859-015-0807-6, NoGOA: predicting noisy GO annotations using evidences and sparse representation, https://doi.org/10.1186/s12859-017-1764-z, Computational discovery of direct associations between GO terms and protein domains, https://doi.org/10.1186/s12859-018-2380-2, Combining learning and constraints for genome-wide protein annotation, https://doi.org/10.1186/s12859-019-2875-5, UNIPred-Web: a web tool for the integration and visualization of biomolecular networks for protein function prediction, https://doi.org/10.1186/s12859-019-2959-2, Interspecies gene function prediction using semantic similarity, https://doi.org/10.1186/s12918-016-0361-5, An expanded evaluation of protein function prediction methods shows an improvement in accuracy, https://doi.org/10.1186/s13059-016-1037-6, The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens, https://doi.org/10.1186/s13059-019-1835-8, Improving Breast Cancer Survival Analysis through Competition-Based Multidimensional Modeling, https://doi.org/10.1371/journal.pcbi.1003047, Machine learning to design integral membrane channelrhodopsins for efficient eukaryotic expression and plasma membrane localization, https://doi.org/10.1371/journal.pcbi.1005786, SVM-Prot 2016: A Web-Server for Machine Learning Prediction of Protein Functional Families from Sequence Irrespective of Similarity, https://doi.org/10.1371/journal.pone.0155290, Predicting protein complexes using a supervised learning method combined with local structural information, https://doi.org/10.1371/journal.pone.0194124, A Survey of Gene Prioritization Tools for Mendelian and Complex Human Diseases, Mining a database of single amplified genomes from Red Sea brine pool extremophilesimproving reliability of gene function prediction using a profile and pattern matching algorithm (PPMA), Learning from Co-expression Networks: Possibilities and Challenges, ProLanGO: Protein Function Prediction Using Neural Machine Translation Based on a Recurrent Neural Network, https://doi.org/10.3390/molecules22101732, Prediction of enzymatic pathways by integrative pathway mapping, Non-H3 CDR template selection in antibody modeling through machine learning, A proposed update for the classification and description of bacterial lipolytic enzymes, Sma3s: A universal tool for easy functional annotation of proteomes and transcriptomes, DeepFunc: A Deep Learning Framework for Accurate Prediction of Protein Functions from Protein Sequences and Interactions, Principles and methods of integrative genomic analyses in cancer, Environmental conditions shape the nature of a minimal bacterial genome, https://doi.org/10.1038/s41467-019-10837-2, Computational identification of protein-protein interactions in model plant proteomes, https://doi.org/10.1038/s41598-019-45072-8, NNTox: Gene Ontology-Based Protein Toxicity Prediction Using Neural Network, https://doi.org/10.1038/s41598-019-54405-6, Protein Secondary Structure Prediction Using Deep Convolutional Neural Fields, FFPred 3: feature-based function prediction for all Gene Ontology domains, Silk fibroin-derived peptide directed silver nanoclusters for cell imaging, The role of ontologies in biological and biomedical research: a functional perspective, Computational analysis and prediction of lysine malonylation sites by exploiting informative features in an integrative machine-learning framework, PANNZER: high-throughput functional annotation of uncharacterized proteins in an error-prone environment, https://doi.org/10.1093/bioinformatics/btu851, Neuro-symbolic representation learning on biological knowledge graphs, https://doi.org/10.1093/bioinformatics/btx275, HFSP: high speed homology-driven function annotation of proteins, https://doi.org/10.1093/bioinformatics/bty262, Phylo-PFP: improved automated protein function prediction using phylogenetic distance of distantly related sequences, https://doi.org/10.1093/bioinformatics/bty704, Prediction of protein group function by iterative classification on functional relevance network, https://doi.org/10.1093/bioinformatics/bty787, DeepGOPlus: improved protein function prediction from sequence, https://doi.org/10.1093/bioinformatics/btz595, Not Only for Egg YolkFunctional and Evolutionary Insights from Expression, Selection, and Structural Analyses of Formica Ant Vitellogenins, DNA-binding protein prediction using plant specific support vector machines: validation and application of a new genome annotation tool, NCBI prokaryotic genome annotation pipeline, Functional sequencing read annotation for high precision microbiome analysis, Importance of metabolic rate to the relationship between the number of genes in a functional category and body size in Peto's paradox for cancer, Neural Network and Random Forest Models in Protein Function Prediction, https://doi.org/10.1109/tcbb.2020.3044230, Sparse Markov chain-based semi-supervised multi-instance multi-label method for protein function prediction, https://doi.org/10.1142/S0219720015430015, Leveraging Physiology for Precision Drug Delivery, https://doi.org/10.1152/physrev.00015.2016, In silico approach to designing rational metagenomic libraries for functional studies, https://doi.org/10.1186/s12859-017-1668-y, CSmetaPred: a consensus method for prediction of catalytic residues, https://doi.org/10.1186/s12859-017-1987-z, Evaluating the impact of topological protein features on the negative examples selection, https://doi.org/10.1186/s12859-018-2385-x, Modeling enzyme-ligand binding in drug discovery, https://doi.org/10.1186/s13321-015-0096-0, PHENOstruct: Prediction of human phenotype ontology terms using heterogeneous data sources, https://doi.org/10.12688/f1000research.6670.1, Recognition of sites of functional specialisation in all known eukaryotic protein kinase families, https://doi.org/10.1371/journal.pcbi.1005975, ProtFus: A Comprehensive Method Characterizing Protein-Protein Interactions of Fusion Proteins, https://doi.org/10.1371/journal.pcbi.1007239, CELLO2GO: A Web Server for Protein subCELlular LOcalization Prediction with Functional Gene Ontology Annotation, https://doi.org/10.1371/journal.pone.0099368, A Multi-Label Supervised Topic Model Conditioned on Arbitrary Features for Gene Function Prediction, Assessing the Performances of Protein Function Prediction Algorithms from the Perspectives of Identification Accuracy and False Discovery Rate, Functional Annotations of Paralogs: A Blessing and a Curse, Review of Biological Network Data and Its Applications, Expansion of the fatty acyl reductase gene family shaped pheromone communication in Hymenoptera, HPO2GO: prediction of human phenotype ontology term associations for proteins using cross ontology annotation co-occurrences, Optimal control nodes in disease-perturbed networks as targets for combination therapy, https://doi.org/10.1038/s41467-019-10215-y, Archetypal transcriptional blocks underpin yeast gene regulation in response to changes in growth conditions, https://doi.org/10.1038/s41598-018-26170-5, DEEPred: Automated Protein Function Prediction with Multi-task Feed-forward Deep Neural Networks, https://doi.org/10.1038/s41598-019-43708-3, Measuring the wisdom of the crowds in network-based gene function inference, https://doi.org/10.1093/bioinformatics/btu715, EGAD: ultra-fast functional analysis of gene networks, https://doi.org/10.1093/bioinformatics/btw695, Predicting multicellular function through multi-layer tissue networks, https://doi.org/10.1093/bioinformatics/btx252, DeepGO: predicting protein functions from sequence and interactions using a deep ontology-aware classifier, https://doi.org/10.1093/bioinformatics/btx624, Ontology-based validation and identification of regulatory phenotypes, https://doi.org/10.1093/bioinformatics/bty605, Candidate gene prioritization with Endeavour, Think globally and solve locally: secondary memory-based network learning for automated multi-species function prediction, Predicting protein function via downward random walks on a gene ontology, https://doi.org/10.1186/s12859-015-0713-y, Trends in genome dynamics among major orders of insects revealed through variations in protein families, https://doi.org/10.1186/s12864-015-1771-2, Gene Ontology Meta Annotator for Plants (GOMAP), https://doi.org/10.1186/s13007-021-00754-1, The evolutionary signal in metagenome phyletic profiles predicts many gene functions, https://doi.org/10.1186/s40168-018-0506-4, Large-scale protein function prediction using heterogeneous ensembles, https://doi.org/10.12688/f1000research.16415.1, Ancient origin of the biosynthesis of lignin precursors, Maize GO Annotation-Methods, Evaluation, and Review (maize-GAMER), Predicting ligand-dependent tumors from multi-dimensional signaling features, https://doi.org/10.1038/s41540-017-0030-3, Data integration and predictive modeling methods for multi-omics datasets, Literature-based automated discovery of tumor suppressor p53 phosphorylation and inhibition by NEK2, A global analysis of function and conservation of catalytic residues in enzymes, https://doi.org/10.1074/jbc.rev119.006289, Formation of Glycated Products and Quality Attributes of Shrimp Patties Affected by Different Cooking Conditions, https://doi.org/10.1080/10498850.2019.1707927, Universal concept signature analysis: genome-wide quantification of new biological and pathological functions of genes and pathways, https://doi.org/10.1093/bioinformatics/btaa763, Extending gene ontology with gene association networks, https://doi.org/10.1093/bioinformatics/btv712, Effusion: prediction of protein function from sequence similarity networks, https://doi.org/10.1093/bioinformatics/bty672, HIPPIE v2.0: enhancing meaningfulness and reliability of proteinprotein interaction networks, Objective risk stratification of prostate cancer using machine learning and radiomics applied to multiparametric magnetic resonance images, Structure to function prediction of hypothetical protein KPN_00953 (Ycbk) from Klebsiella pneumoniae MGH 78578 highlights possible role in cell wall metabolism, Learning a functional grammar of protein domains using natural language word embedding techniques, Analysis of protein targets in pathogenhost interaction in infectious diseases: a case study on Plasmodium falciparum and Homo sapiens interaction network, The ortholog conjecture revisited: the value of orthologs and paralogs in function prediction, https://doi.org/10.1093/bioinformatics/btaa468, Towards CRISPR/Cas crops - bringing together genomics and genome editing, Systematic Analysis of Challenge-Driven Improvements in Molecular Prognostic Models for Breast Cancer, https://doi.org/10.1126/scitranslmed.3006112, A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction, Predicting human protein function with multi-task deep neural networks, https://doi.org/10.1371/journal.pone.0198216, PANDA: Protein function prediction using domain architecture and affinity propagation, https://doi.org/10.1038/s41598-018-21849-1, https://doi.org/10.1038/s41598-018-38381-x, Leveraging implicit knowledge in neural networks for functional dissection and engineering of proteins, https://doi.org/10.1038/s42256-019-0049-9, Protein secondary structure prediction using deep convolutional neural fields, https://doi.org/10.48550/arxiv.1512.00843, https://doi.org/10.48550/arxiv.1601.00891, Report on the 2011 Critical Assessment of Function Annotation (CAFA) meeting, Use of Modern Chemical Protein Synthesis and Advanced Fluorescent Assay Techniques to Experimentally Validate the Functional Annotation of Microbial Genomes, Breaking the High-Throughput Bottleneck: New tools help biologists integrate complex datasets, Using support vector machine for improving protein-protein interaction prediction utilizing domain interactions, Indiana Univ., Bloomington, IN (United States).